Workflows¶
MIAAIM contains 4 major workflows:
1. High-Dimensional Image Preparation with the HDIprep workflow
2. High-Dimensional Image Registration with the HDIreg workflow
3. Tissue state modelling with the PatchMAP workflow and
4. Cross-system/tissue information transfer with the i-PatchMAP workflow.
For a comprehensive table of input options for these workflows, see the Parameter Reference section. Up-to-date input options and docker containerized versions of each workflow are available on GitHub:
Workflow |
Resource |
|---|---|
hdiprep |
|
hdireg |
|
patchmap/i-patchmap |
Image Preparation (HDIprep)¶
Image preparation begins the workflow for MIAAIM, and it is designed to provide processed images to the HDIreg image registration workflow. HDIprep can process high-parameter images as well as histological stains.
HDIprep works by chaining together multiple operations in sequence to process images. Each operation is indicated as an argument in a YAML parameter file that is passed to the HDIprep workflow.
Tip
If your images have more than 3 channels, you should apply HDIprep as indicated in the High Dimensional Image Processing section. If your images have 3 or less channels, you should apply HDIprep as indicated in the Histological Image Processing section.
Warning
All HDIprep parameter files should end with the ExportNifti1 function
so that an output is produced!
YAML Parameter File Input Basics¶
HDIprep parses YAML parameter files as Python would. Input to the HDIprep workflow
requires two sections to be completed in YAML parameter files for both a fixed
image, which will remain unmoved during image registration, and a moving image,
which will be transformed. The sections that must be completed are the ProcessingSteps
and InputOptions sections. Below is an example parameter file for
image compression with HDIprep:
ImportOptions: # indicates import options
flatten: True # flatten the image for dimension reduction
subsample: True # subsample pixels
method: 'grid' # method to use for subsampling
grid_spacing: (5,5) # subsampling grid spacing
ProcessingSteps: # indicates processing steps
- RunOptimalUMAP: # steady state UMAP compression
n_neighbors: 15 # nearest neighbors for UMAP
metric: 'euclidean' # metric for UMAP
random_state: 1221 # random state for UMAP reproducibility
- SpatiallyMapUMAP # spatial reconstruction of UMAP embedding
- ExportNifti1 # export processed image as NIfTI
1. ImportOptions parameters are indicated as key-value pairs
with a colon (:) For example,
the above snippet will import high-dimensional image data, it will flatten it
into an array with the flatten: True, subsample the flattened array with
subsample: True using a uniformly spaced grid with grid spacing of 5 pixels.
2. ProcessingSteps in HDIprep are indicated
with the - flag. These steps will be run sequentially. Parameters within each
processing step are indicated as key-value pairs with a colon (:) For example,
the above snippet will run steady state UMAP compression using the RunOptimalUMAP
function with the given n_neighbors, metric, and random_state parameters
specified.
Then, pixels in the UMAP embedding will be mapped to their respective spatial location
on the tissue with using the SpatiallyMapUMAP function. This reconstructed
UMAP compressed image will then be
exported as an image with the ExportNifti1 function.
Note
MIAAIM expects the NIfTI format (.nii suffix) as input for the HDIreg
workflow. HDIprep therefore
exports processed images using the ExportNifti1
function. NIfTI provides memory-mapping capabilities within Python
that save RAM in HDIreg. However, OME-TIF, HDF5, imzML, and NIfTI can be used as input
formats for HDIprep!
High-Parameter Image Processing¶
MIAAIM processes high-parameter images using a newly developed image compression method. This method is based off of UMAP. The HDIprep workflow adds functionality to subsample images for rapid UMAP compression, and it can embed data in a dimensionality that preserves data information while minimizing the necessary dimensionality of the embedding space (number of channels in the compressed image).
Implementing this new compression method is indicated in the HDIprep YAML parameter
file under ProcessingSteps as RunOptimalUMAP. After an embedding
is created with RunOptimalUMAP, map the pixels back to their spatial location
with SpatiallyMapUMAP.
Note
The HDIprep workflow can also apply a neural network variant of UMAP to
scale to very large images. Implement this version by using the function
RunOptimalParametricUMAP.
Tip
The HDIprep workflow can accept a mask in the TIFF format to focus dimension
reduction on a region of interest on a larger section. This could be useful
for very large noisy images. Use this feature by specifying
mask: True under ImportOptions. Make sure the mask is
in the input directory with your image data and be sure to name it with your
image’s name followed by the suffix -mask. For example, the mask associated
with a a moving image named moving.nii should be named moving-mask.tif.
See this in action below:
Histological Image Processing¶
MIAAIM supports parallelized image smoothing and morphological operations, such as thresholding to create masks, opening, closing, and filling for histological image preprocessing. These are typically applied as sequential image processing options.
HDIprep Implementation Guide¶
Image Data Type |
Suggested HDIprep YAML File Contents |
|---|---|
High-Dimensional (>3 channels) |
ImportOptions:
flatten: True
subsample: True
method: 'grid'
grid_spacing: (3,3)
ProcessingSteps:
- RunOptimalUMAP:
n_neighbors: 15
metric: 'euclidean'
landmarks: 3000
dim_range: (1,10)
random_state: 1221
- SpatiallyMapUMAP
- ExportNifti1
|
Low-Dimensional (<=3 channels) |
ImportOptions:
flatten: False
subsample: None
mask: True # True if you have a custom input mask, otherwise set to False
ProcessingSteps:
- RunOptimalUMAP:
n_neighbors: 15
metric: 'euclidean'
landmarks: 3000
dim_range: (1,10)
random_state: 1221
- SpatiallyMapUMAP
- ExportNifti1
ProcessingSteps:
- MedianFilter:
filter_size: 25 # median filtering
parallel: True
- Threshold:
type: 'otsu' # create mask with thresholding
- Open:
disk_size: 20 # morphological opening
parallel: True
- Close:
disk_size: 40 # morphological closing
parallel: True
- Fill # fill holes in mask
- Open:
disk_size: 15 # morphological opening
parallel: True
- ApplyManualMask # apply manual input mask
- NonzeroBox # perform nonzero slicing for constant padding
- ApplyMask
- ExportNifti1 # export as nifti
|
Note
The above processing steps are based on histological images scaled to a resolution of ~1.2 micron/pixel.
Image Registration (HDIreg)¶
Image registration in MIAAIM is performed in the HDIreg workflow. Like the HDIprep workflow, HDIreg can process high-parameter images as well as histological stains.
HDIreg works by supplying an Elastix parameter file in a pairwise registration procedure between a fixed image (reference image), and a moving image.
Tip
If your images have more than 3 channels, you should apply HDIprep as indicated in the High Dimensional Image Processing section. If your images have 3 or less channels, you should apply HDIprep as indicated in the Histological Image Processing section.
Warning
All HDIprep parameter files should end with the ExportNifti1 function
so that an output is produced!
HDIreg Implementation Guide¶
Image Data Type |
Suggested HDIreg Elastix Text File Contents |
|---|---|
Mixed High (>3) and Low (<= 3 channels) Dimensional Images |
Registration “MultiResolutionRegistrationWithFeatures” Metric “KNNGraphAlphaMutualInformation” ImageSampler “MultiInputRandomCoordinate” |
High (>3 channels) Dimensional Images |
Registration “MultiResolutionRegistrationWithFeatures” Metric “KNNGraphAlphaMutualInformation” ImageSampler “MultiInputRandomCoordinate” |
Low (<=3 channels) Dimensional Images |
Registration “MultiResolutionRegistration” Metric “AdvancedMattesMutualInformation” ImageSampler “RandomCoordinate” |
Tissue State Modelling (PatchMAP)¶
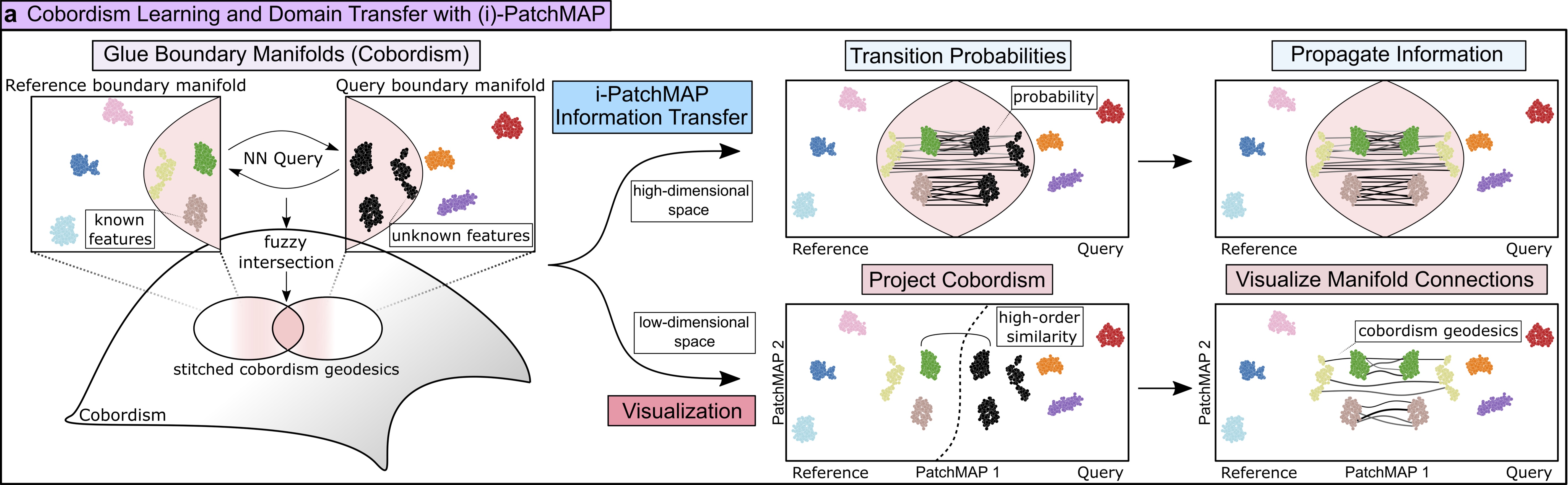
Continuity between tissue state in MIAAIM is performed with the PatchMAP workflow. PatchMAP explicitly constructs a manifold that captures structure between data manifolds.
PatchMAP is currently implemented in the Python implementation of MIAAIM. See the Python workflows for an example use case.
Cross-System/Tissue Information Transfer (i-PatchMAP)¶
i-PatchMAP utilizes the manifold constructed between data separate data manifolds in the PatchMAP workflow to transfer information from a reference to a query data set.